Announcing 2022 Translational Health Data Science Fellows
We are excited to announce the 2022 cohort for the inaugural Translational Health Data Science fellowship program. A goal of this program is to provide advanced master’s and doctoral students with the opportunity to participate in mentored research that advances their graduate progress and contributes to the development and/or implementation of data science applications to the human health sciences. This fellowship is open to students from interdisciplinary fields ranging from computer science, engineering, social sciences, biomedicine, informatics, to public health and beyond. This 9-month fellowship program is supported by CITRIS, CTSC, and the Department of Public Health Sciences in collaboration with DataLab.
2022 Fellows

Charlie Fornaca: “Enabling Synthetic Data Usage for Medical Research”
Mentors: Dr. Vladimir Filkov (Computer Science) and Dr. Nick Anderson (Health Informatics)
Acquiring data can be a major hurdle to any data science problem. Sometimes there isn’t enough data or, as is particularly the case for healthcare data, it may contain sensitive information such as personal identifiers that should not be shared. By generating synthetic health data, we aim to overcome obstacles of data access and privacy concerns and thereby allow for quicker and broader use of data by the research community. Charlie’s research explores and compares different efforts and methods for generating synthetic healthcare data, with a goal of proposing workflows and platforms for supporting health researchers at UC Davis.
Charlie is pursuing her master’s degree in Computer Science at UC Davis. Her interests include applied data science, AI & machine learning, and accessibility technology. After graduation she plans to work as a data scientist serving the public interest.
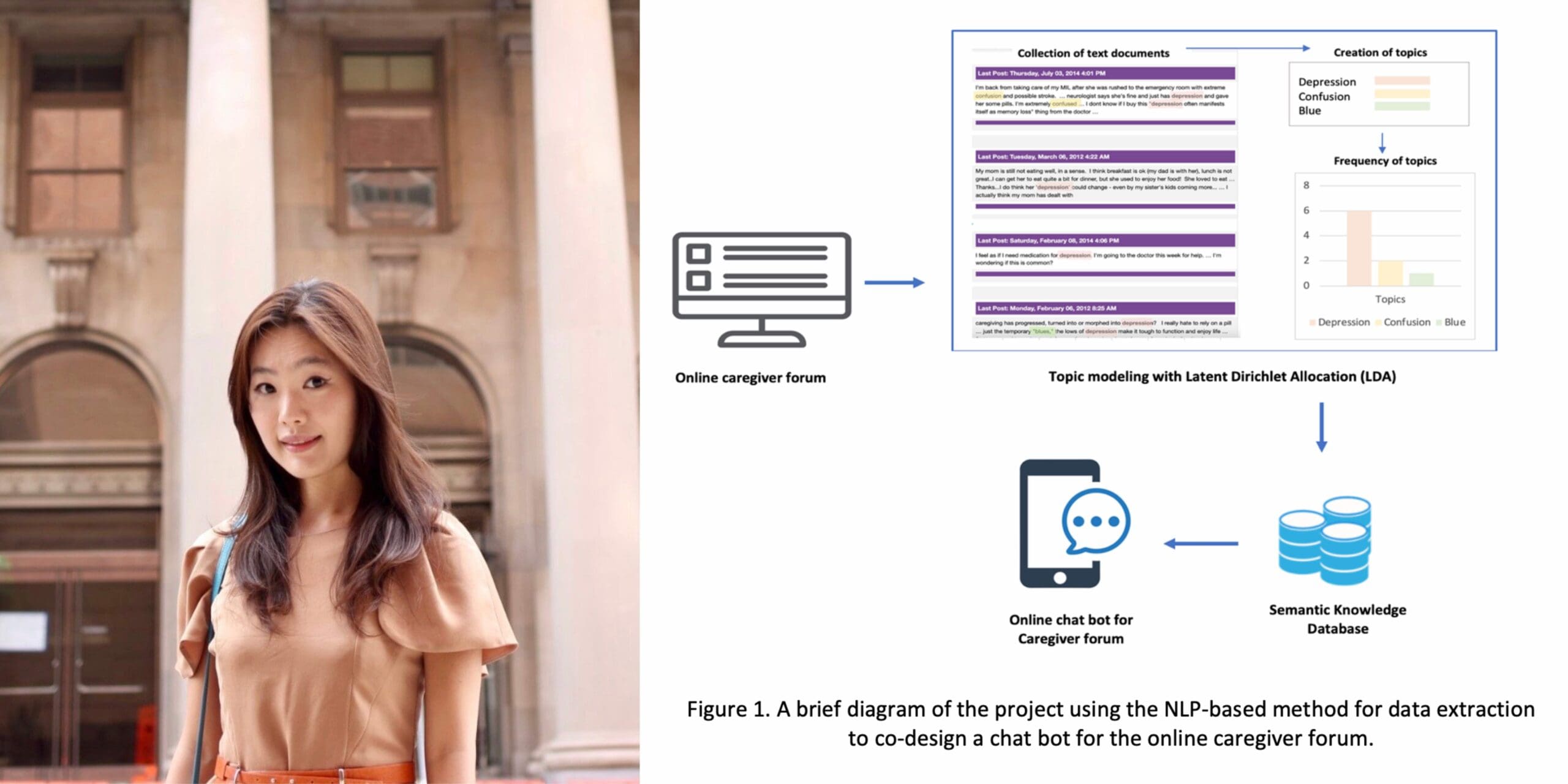
Jiyeong Kim: “Helping Caregivers find Tailored Mental Health Information by Applying NLP to Online Caregiver Forums”
Mentors: Dr. Yong Choi (Public Health Sciences) and Dr. Vladimir Filkov (Computer Science)
Relatives and other informal caregivers of Alzheimer’s disease and related dementia patients are at risk for mental distress, but many do not know where to seek help for their own mental health. Jiyeong’s research applies machine learning and natural language processing methods to the discourse on active web-based caregiver forums to programmatically determine common and evolving caregiver mental health needs. This approach allows for validation of human-based qualitatively assessed caregiver needs data, as well as the capture of new data. Extensions of this work include the development of potential intervention strategies, including AI-driven conversational agents that can be embedded into such online forums to provide caregivers with just in time resources for their specific mental health needs.
Jiyeong Kim is a second-year Ph.D. student in Public Health Sciences. Her healthcare background spans from working as a cancer center clinical pharmacist, medical representative for Johnson and Johnson, founder and CEO of a health education app start-up, and the COO of a women’s health e-commerce company. Her research harnesses digital health technologies to tackle mental health issues in vulnerable populations.
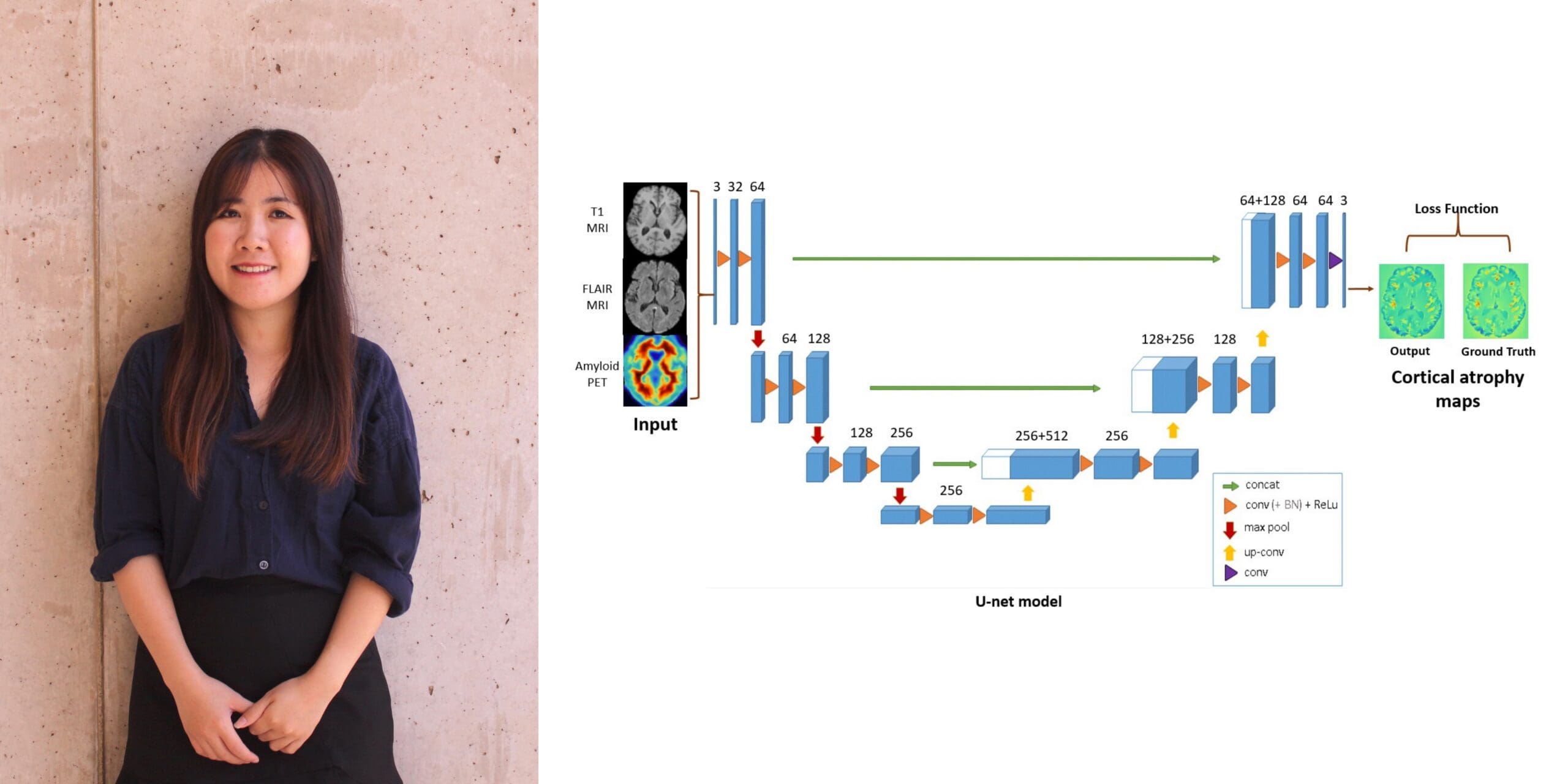
Linh Nguyen Ngoc Le: “Predicting Future Atrophy Maps for Early Detection of Alzheimer’s Disease”
Mentors: Dr. Audrey Fan (Biomedical Engineering) and Dr. Jinyi Qi (Biomedical Engineering)
Alzheimer’s disease is the most common form of dementia and can impose a great social and economic burden on both patients and their families. To aid in early-stage detection of Alzheimer’s disease progression, Linh is developing and applying a multi-modal neural network to medical images to predict future longitudinal brain atrophy in elderly patients at risk of cognitive decline.
Linh is a Ph.D. student in Biomedical Engineering interested in using advanced imaging algorithms and quantitative imaging biomarkers to improve the diagnosis of neurodegenerative diseases. Linh graduated from University of California, San Diego with a B.S. in Bioengineering and worked in a computational neurology research group at the Salk Institute for Biological Studies. Post-graduation, Linh worked for Cortechs.ai as part of a project exploring the development of a neuroimaging software product which automatically segments MRI and PET scan images.
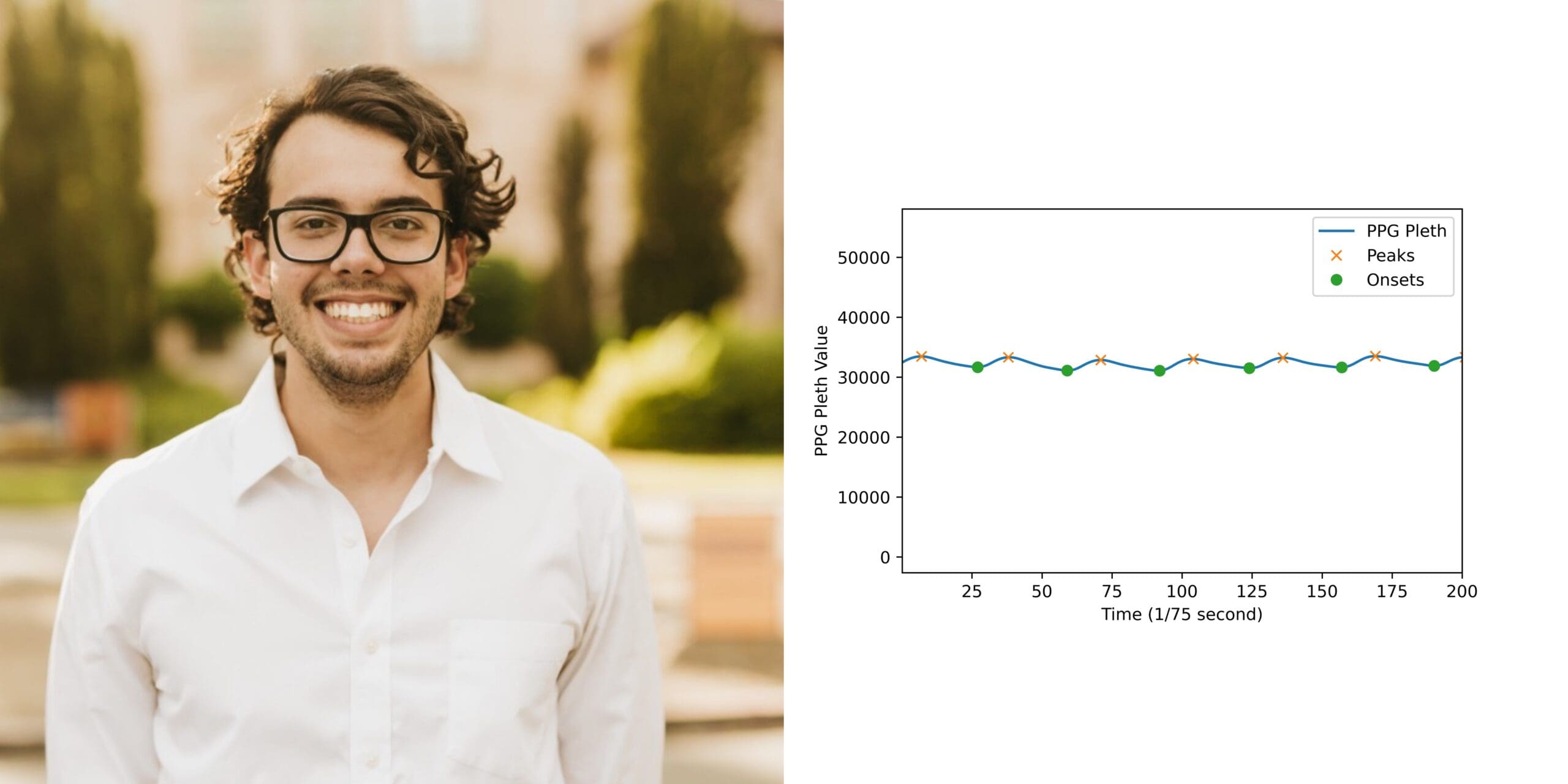
Luca Cerny Oliveira: “Deep Semi-Supervised Learning for Reduced Labile Use in 1D Medical Signals”
Mentors: Dr. Chen-Nee Chuah (Electrical and Computer Engineering) and Dr. Heather Siefkes (Pediatrics)
Deep learning has great potential for applications to predictive health applications. While these algorithms often use supervised learning and can yield high accuracy, this strategy can be very resource-intensive and require clinicians to label and annotate large amounts of data. Semi-supervised learning can achieve similar results to supervised learning while utilizing significantly less annotated data. Luca’s research investigates the use of semi-supervised learning on one-dimensional medical signals such as ventilator waveforms, electrocardiograms, and photoplethysmography images.
Luca is a master’s student in Electrical and Computer Engineering from Brazil whose work focuses on machine learning and the development of smart health technologies that can aid in predictive diagnosis and pathology research.

Ellen Osborn: “Leveraging Computational Tools for Genome Analysis of a Human Pathogenic Gene”
Mentors: Dr. Suma Shankar (Pediatrics) and Dr. John McPherson (Biochemistry and Molecular Medicine)
More than 100 genes are associated with autism spectrum disorder (ASD), and many individuals with ASD harbor rare pathogenic variants. Molecular diagnoses of pathogenic variants can aid in both clinical care and our overall understanding of ASD. However, due to underdeveloped computational approaches in variant interpretation as well as the use of study cohorts without clear phenotypic delinations, modern genome sequencing technologies do not always identify these variants in a clinical setting. Ellen’s research addresses this by developing a bioinformatic pipeline to identify likely pathogenic variants from whole genome sequences. By identifying novel variants and genes associated with ASD in a large non-verbal ASD study cohort, Ellen hopes to increase understanding of the genetic etiology of ASD and contribute to improved patient clinical care.
Ellen is a M.S. student in Integrative Genetics and Genomics whose primary research interest is the clinical interpretation of human genomic variation, particularly involving rare disease. Ellen’s goal is to earn an M.D./Ph.D. and become a medical geneticist working directly with patients, researching rare genetic diseases, and teaching genetics to undergraduates.

Olivia Sattayapiwat: “Advancing Evidence for Associations Between Benign Breast Diagnoses and Future Breast Cancer Risk”
Mentors: Dr. Diana Miglioretti (Public Health Sciences) and Dr. Theresa Keegan (Internal Medicine)
Breast biopsies commonly reveal benign breast diseases. While not indicative of breast cancer, some of these benign diagnoses are known to be associated with high risk for future development of the disease. Olivia’s research applies supervised machine learning methods to longitudinal data gathered from breast imaging registries across the United States to identify specific breast disease diagnoses and accurately quantify their associations with breast cancer risk.
Olivia Sattayapiwat is a Ph.D. candidate in Epidemiology. Her research interests lie mainly in cancer epidemiology, working to improve identification and measurement of risk factors to build better cancer risk prediction models and targeted intervention strategies. Prior to joining UC Davis, Olivia received her B.S. in Molecular Cell Biology from UC Berkeley, her M.P.H. in Environmental Health Sciences from UCLA, and her M.S. degree in Epidemiology from USC. Between earning her master’s degrees, Olivia worked as a data analyst at the Breast Diagnostic Center at Torrance Memorial Medical Center and as a research associate at Kaiser Permanente Southern California, supporting cancer epidemiology studies related to prostate, non-Hodgkin’s lymphoma, adolescent and young adult, bladder, ovarian, and breast cancer.
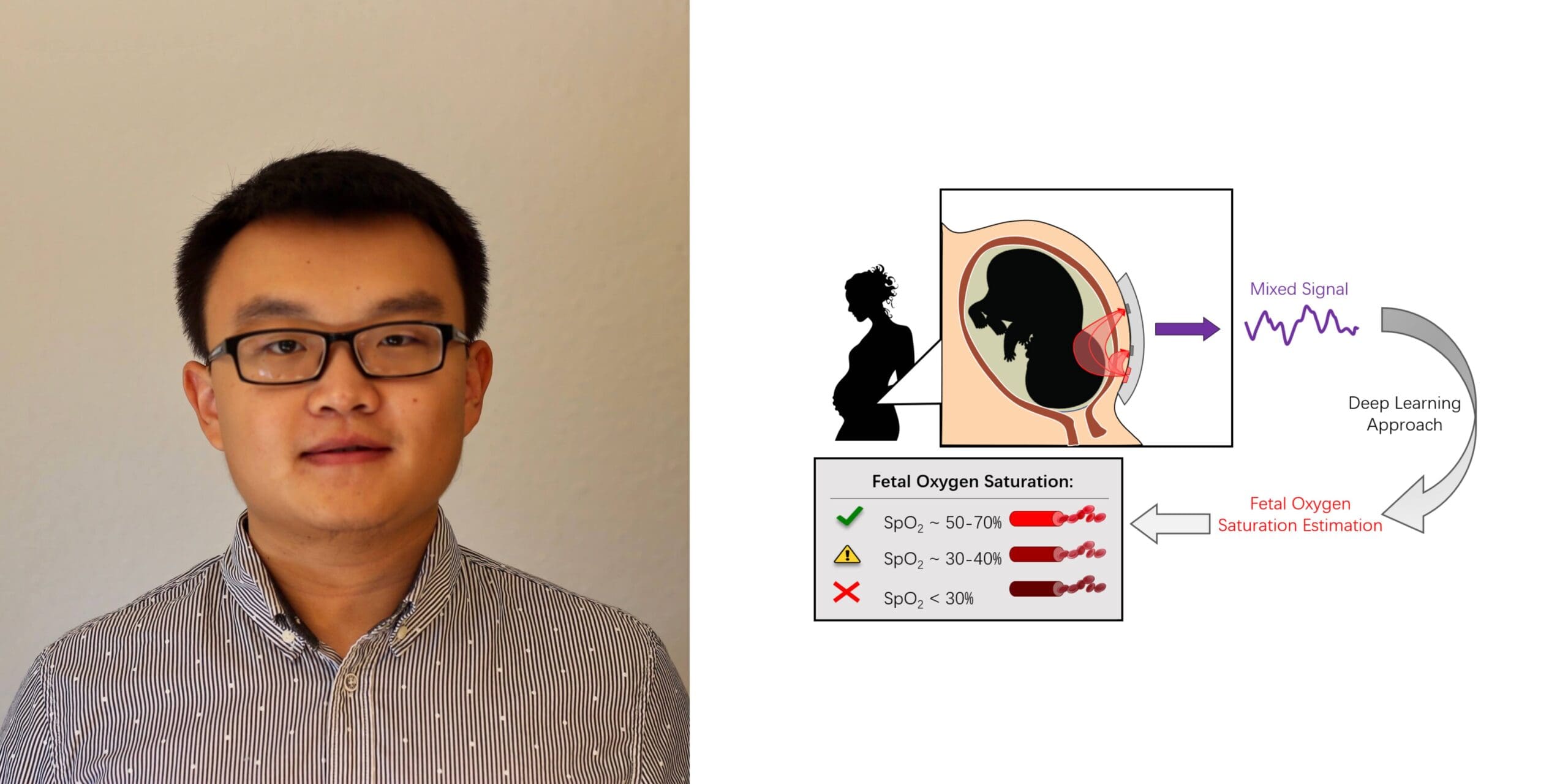
Weitai Qian: “Enhanced Intrapartum Fetal Monitoring via Context-Aware Data Integration”
Mentors: Dr. Soheil Ghiasi (Electrical and Computer Engineering) and Dr. Herman Hedriana (Obstetrics and Gynecology)
Most obstetricians utilize cardiotocography (CTG) to assess signs of fetal distress during childbirth, but this technique is relatively unreliable in measuring oxygen supply. Insufficient oxygen supply can cause birth asphyxia or hypoxic ischemic encephalopathy, with severe adverse outcomes. Weitai’s research focuses on the potential of a new technique, transabdominal fetal oximetry, which applies deep learning technologies to maternal measurements to estimate fetal oxygen saturation levels and more accurately assess the well-being of a baby during childbirth. By developing generative adversarial networks to augment limited data from animal and human trials, this research aims to increase the accuracy of transabdominal fetal oximetry derived assessments of fetal oxygen saturation, thereby decreasing in-labor risks for both babies and birthing parents during labor and delivery.
Weitai Qian is a third-year Ph.D. student in Electrical and Computer Engineering. He received his B.S. in Computer Engineering from University of Wisconsin-Madison. His research interests center around biomedical applications including health data processing and hardware implementation.